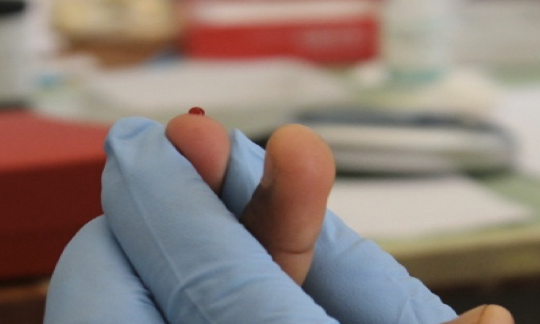
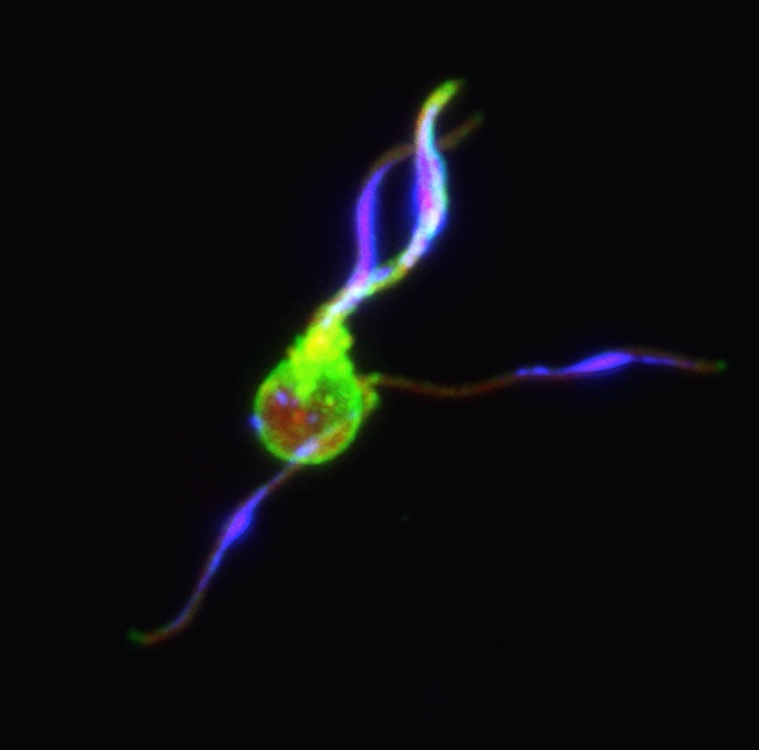
In malaria parasites, the disease is caused by asexual parasites, but sexual forms are the only ones capable of transmitting the infection to mosquitoes. Developmental events leading to differentiation into either a male or a female form are poorly understood. In this project, using sophisticated single cell technology, we will find and study genes involved in the sex determining pathway. We will identify host cues which influence commitment to the male or female lineage in naturally infected carriers in Cameroon. The findings of this project will provide biological insights into malaria sexual behaviour and potentially lead to identification of new drug targets. Targeting sex determination could potentially sterilise parasites and interrupt transmission.
CURRENT TEAM MEMBERS

Interested in parasite and host factors influencing gametocyte carriage and infectivity to mosquitoes.
Co-supervised with Isa Morlais

Interested in sex determination of Plasmodium and factors influencing infectivity to mosquitoes

Studying sexual differentiation of Plasmodium. Co-supervised with Stephane Ranque (IHU, Marseille)

Investigating malaria parasites in the lab & the field. Founder of Scicling, a public engagement initiative (www.scicling.org)

Interested in Plasmodium molecular and cellular biology underlying drug resistance and DNA replication

Interested in Plasmodium malariae and invasion. Co-supervised with Laurent Dembele (USTTB, Bamako)
CURRENT PROJECTS
click for summary ...
Collaboration between Fred Ariey (PI), Robert Menard (co-I), Jerome Clain (co-I) and Arthur Talman (co-I)
click for summary ...
Artemisinin derivatives (ART) are currently the last approved anti-malarial drug against which resistance has not yet spread widely. Worryingly, since 2008 the efficacy of ART-base combination therapies has decreased in South-East Asia, due to the emergence and spread of ART resistance (ARTR). Spread of ARTR to sub-Saharan Africa, where most clinical cases and deaths occur, would be catastrophic and threaten the world’s malaria control and elimination efforts. ARTR was first defined by the delayed clearance time of P. falciparum parasites from the peripheral blood of Cambodian patients treated with ART monotherapy, associated with an increase in treatment failures. Strikingly, these ‘resistant’ parasites were fully or only slightly less sensitive to ART in vitro, as defined by standard growth inhibition assays under continuous drug pressure. Delayed parasite clearance in patients, however, correlated with the ring-stage survival assay (RSA), which measures the percentage of early ring-stage parasites surviving a clinically relevant 6 h-pulse of DHA. Parasites ‘resisting’ a first RSA display a similar survival rate when subjected to a second RSA. Clearly, such resistance does not respond to a classical resistance phenotype. Rather, they suggested that parasites subjected to ART at early developmental stages (rings) could escape aggression by entering in some form of dormancy or cell cycle dysregulation. The first and currently only ARTR markers are all mutations that cluster in the Kelch domain of the K13 protein. These K13 mutations associate with delayed parasite clearance in patients across South-East Asia. The K13 C580Y substitution – accounting for 80% of resistant cases – was shown to be sufficient for conferring resistance in Cambodian and laboratory P. falciparum strains using genome-editing technology. Our understanding of ARTR is still hampered by the lack of a formal investigation of activation and nature of cell cycle modulation/dormancy pathway(s). In this collaborative project, our group will participate by harnessing the power of single cell technology to understand the nature of dormancy. A deep understanding of the cellular and molecular basis of resistance is central to our capacity to fully circumvent the issue of recrudescence, and better grasp how P. falciparum adapts its growth to hostile environments such as drug-induced oxidative stress.
RECENT PUBLICATIONS
Science
click for abstract ...
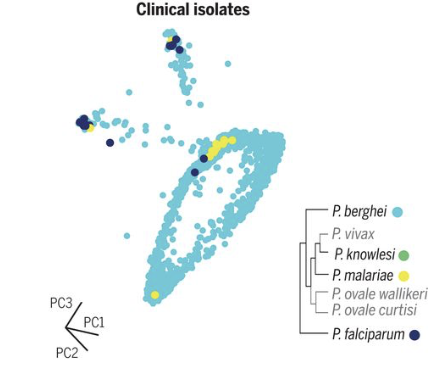
Malaria parasites adopt a remarkable variety of morphological life stages as they transition through multiple mammalian host and mosquito vector environments. We profiled the single-cell transcriptomes of thousands of individual parasites, deriving the first high-resolution transcriptional atlas of the entire Plasmodium berghei life cycle. We then used our atlas to precisely define developmental stages of single cells from three different human malaria parasite species, including parasites isolated directly from infected individuals. The Malaria Cell Atlas provides both a comprehensive view of gene usage in a eukaryotic parasite and an open-access reference dataset for the study of malaria parasites.
Read more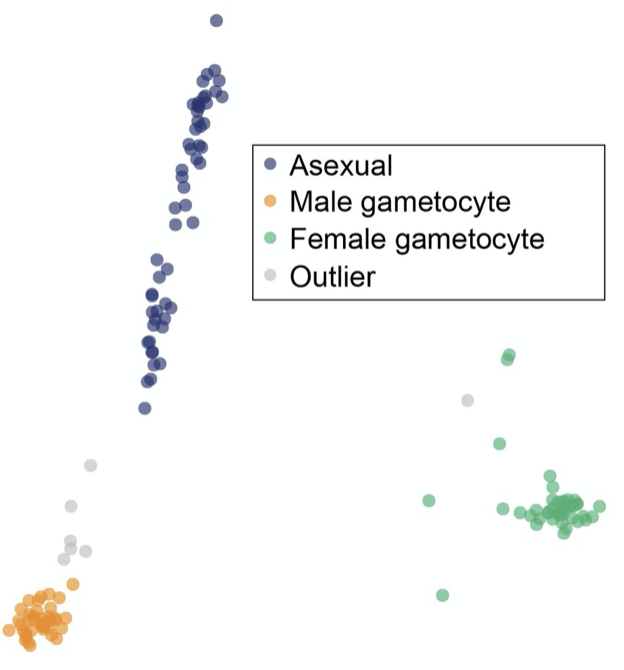
Elife
click for abstract ...
Single-cell RNA-sequencing is revolutionising our understanding of seemingly homogeneous cell populations but has not yet been widely applied to single-celled organisms. Transcriptional variation in unicellular malaria parasites from the Plasmodium genus is associated with critical phenotypes including red blood cell invasion and immune evasion, yet transcriptional variation at an individual parasite level has not been examined in depth. Here, we describe the adaptation of a single-cell RNA-sequencing (scRNA-seq) protocol to deconvolute transcriptional variation for more than 500 individual parasites of both rodent and human malaria comprising asexual and sexual life-cycle stages. We uncover previously hidden discrete transcriptional signatures during the pathogenic part of the life cycle, suggesting that expression over development is not as continuous as commonly thought. In transmission stages, we find novel, sex-specific roles for differential expression of contingency gene families that are usually associated with immune evasion and pathogenesis.
Read moreNEWS

Scicling is a Public Engagement initiative conceived by Dr. Alejandro Marín-Menéndez (Alex), to bring cutting-edge Science to Secondary Schools, and the general public, around the world. As you might have guessed rightly, he also aims to do so in a sustainable way: cycling!
more ...
The main aims are to show that: a) Science is in every aspect of our lives. Focusing on how DNA-based technologies are reshaping our understanding of infectious diseases; b) Science can be applied anywhere and, in many cases, in a simple way; and c) Science is fun! Last year, Alex cycled across the Canary Islands and Uruguay and visited over 25 Secondary Schools engaging with more than 1000 students and their communities. After an uncertain year and adapting to the current circumstances, on August 21st a new action commenced and instead of bringing ‘Science to Schools’, is bringing ‘Scientists to Homes’. Follow it on Instagram and/or Twitter (@Scicling) or visit the website (www.scicling.org) to keep you updated.
Read more
After a Master II in the lab, Emilie is sticking around to continue deciphering dormancy and DNA replication in Plasmodium.
PHOTO GALLERY
Montpellier